Capturing fine-scale structure
Biologically, a population is a group of living organisms of the same species that are living together and have the capability of interbreeding. Evolutionary processes such as migration, natural selection etc., has overtime contributed to the variations in the gene makeup of individuals that makes up a particular population. Subpopulations are as a result of these variations that are present in genes of individuals of a population. Population studies have several importance among which are; Information gotten from population studies can be used to infer population history, also studying the population of a place can reveal predisposing factors of disease in that place and several others. Population stratification is one of several population studies that is being carried out. Population stratification is the difference in allele frequencies between subpopulations in a population, possibly due to different ancestry, especially in the context of association studies. Analysis of population stratification must meet 4 main challenges namely:
- Detecting the structure of a population
- Assigning individuals to subpopulations
- Determining the number of optimal or primal subpopulations( i.e the dominant subpopulations)
- Determining the proportion of ancestral subpopulation.
Overtime there has been an immense increase in the numbers and types of genotypes that exist among populations and this increase has resulted in difficulties in carrying out certain populations studies such as correctly estimating the subpopulations and assigning individuals to them, the increase has also resulted in difficulties in carrying out population structure analysis. Principal components analysis (PCA) is the common computational method used in carrying out population structure analysis. It can accurately detect population structure but has limitations in its accuracy in resolving subpopulations and assigning individuals to them due to the increasing numbers of genotype as time progresses with high-throughput genotyping technology. Therefore there is need for a more efficient computational method which despite the constant addition of genotypes to the population would accurately resolve subpopulations and assign individuals to them.
A population structure analysis algorithm has been developed which does not have the limitations present in the principal component analysis (PCA). This more efficient algorithm is called iterative pruning PCA (ipPCA). It is capable of resolving subpopulations and assigning individuals to them.
This new computational method is highly efficient and is not limited by the constant influx of genotypes into the population. Therefore when carrying out population structure analysis, it is advisable to use iterative pruning PCA (ipPCA) method since it does not have the limitations present in the common principal components analysis (PCA).
Read the technical details at DOI: 10.1186/s13029-019-0072-6
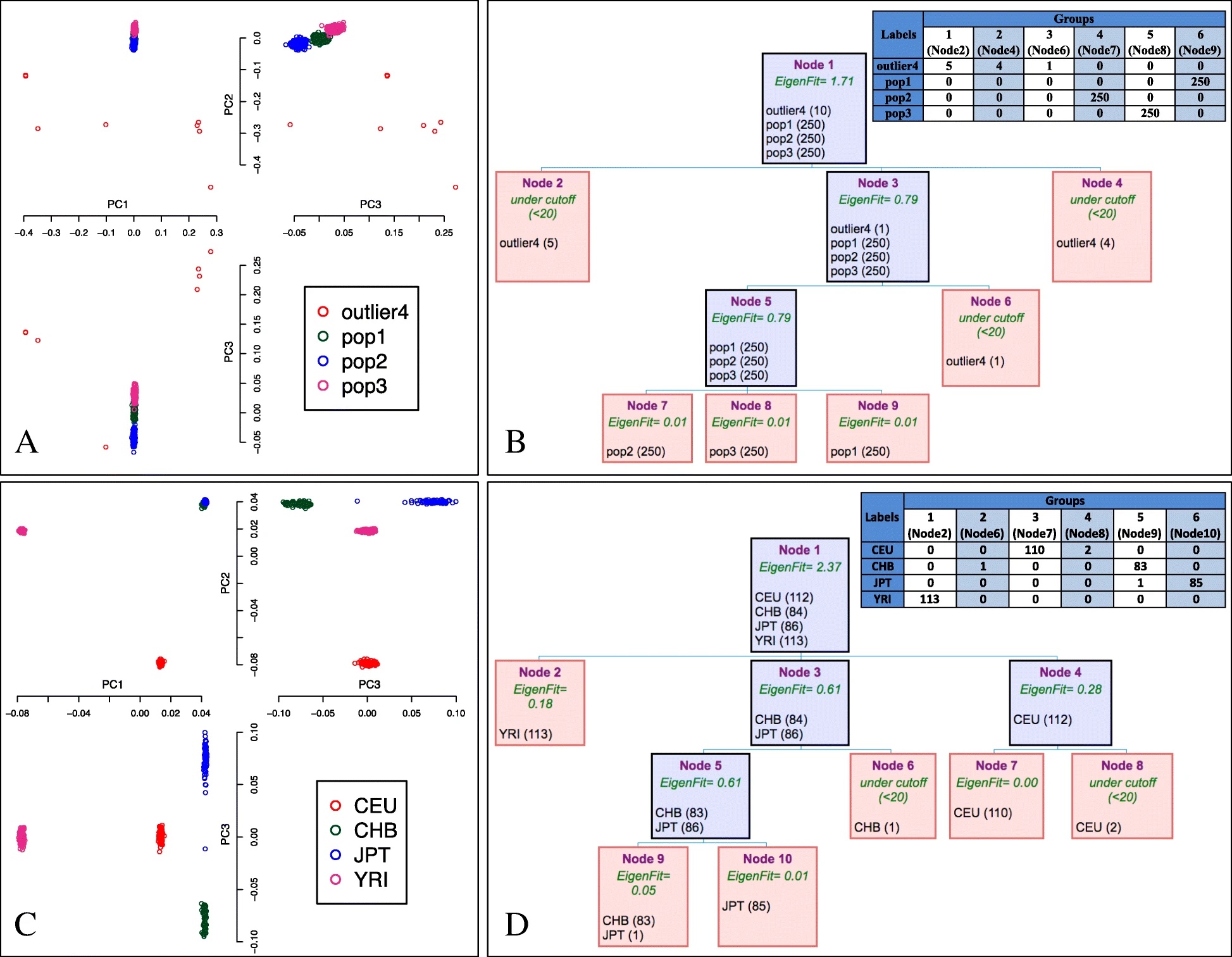 Preview figure from the article
Preview figure from the article