KRIS (Keen and Reliable Interface Subroutines for bioinformatic analysis) is the package providing useful functions which are needed for bioinformatic analysis such as calculating linear principal components from numeric data and Single-nucleotide polymorphism (SNP) dataset, calculating fixation index (Fst) using Hudson method, creating scatter plots in 3 views, handling with PLINK binary file format, detecting rough structures and outliers using unsupervised clustering, and calculating matrix multiplication in the faster way for big data..
Installation
You can install the released version of KRIS from CRAN with:
install.packages("KRIS")Alternatively, you can install the dev version of KRIS from Gitlab with
install.packages("remotes")
remotes::install_github("kridsadakorn/kris", dependencies = TRUE)Document
You can see the reference manual from: https://www.biostatgen.org/kris/
Example
This is a basic example which shows you how to use the package:
example_SNP consists of the simulated SNP data simsnp and the sample labels sample_labels.
summary(simsnp)
#> Length Class Mode
#> snp 206000 -none- numeric
#> snp.info 6 data.frame list
#> ind.info 6 data.frame list
summary(sample_labels)
#> Length Class Mode
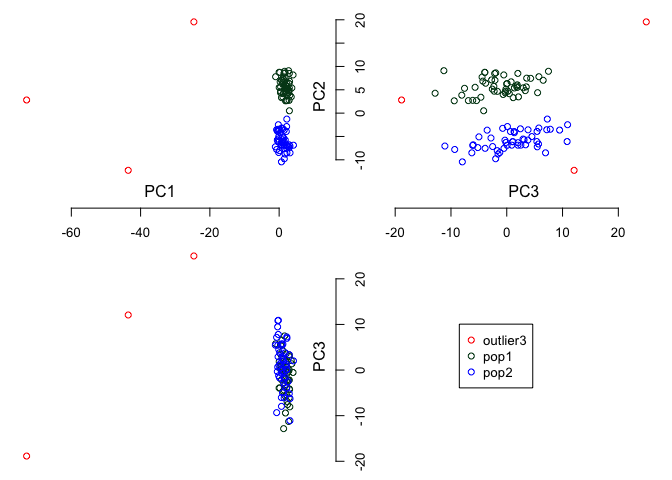
#> 103 character characterTo calcualte the first three linear principal components from SNP data:
PCs <- cal.pc.linear(simsnp$snp, no.pc = 3)You can visualize 3 dimensions for the first three principal components as: